Fitting TESS data#
import exoplanet
exoplanet.utils.docs_setup()
print(f"exoplanet.__version__ = '{exoplanet.__version__}'")
exoplanet.__version__ = '0.5.4.dev27+g75d7fcc'
In this tutorial, we will reproduce the fits to the transiting planet in the Pi Mensae system discovered by Huang et al. (2018). The data processing and model are similar to the Joint RV & transit fits case study, but with a few extra bits like aperture selection and de-trending.
To start, we need to download the target pixel file:
import numpy as np
import lightkurve as lk
import matplotlib.pyplot as plt
from astropy.io import fits
lc_file = lk.search_lightcurve(
"TIC 261136679", sector=1, author="SPOC"
).download(quality_bitmask="hardest", flux_column="pdcsap_flux")
lc = lc_file.remove_nans().normalize().remove_outliers()
time = lc.time.value
flux = lc.flux
# For the purposes of this example, we'll discard some of the data
m = (lc.quality == 0) & (
np.random.default_rng(261136679).uniform(size=len(time)) < 0.3
)
with fits.open(lc_file.filename) as hdu:
hdr = hdu[1].header
texp = hdr["FRAMETIM"] * hdr["NUM_FRM"]
texp /= 60.0 * 60.0 * 24.0
ref_time = 0.5 * (np.min(time) + np.max(time))
x = np.ascontiguousarray(time[m] - ref_time, dtype=np.float64)
y = np.ascontiguousarray(1e3 * (flux[m] - 1.0), dtype=np.float64)
plt.plot(x, y, ".k")
plt.xlabel("time [days]")
plt.ylabel("relative flux [ppt]")
_ = plt.xlim(x.min(), x.max())
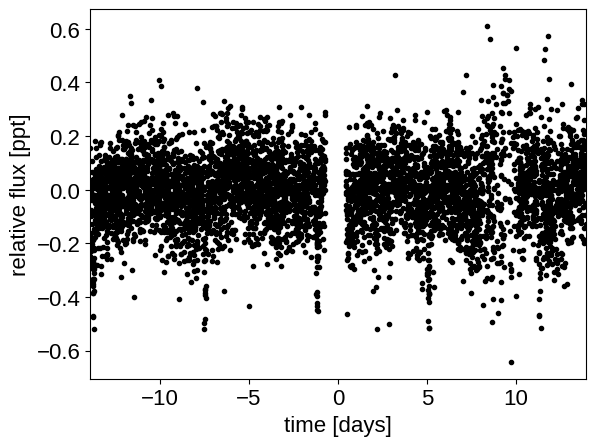
Transit search#
Now, let’s use the box least squares periodogram from AstroPy (Note: you’ll need AstroPy v3.1 or more recent to use this feature) to estimate the period, phase, and depth of the transit.
from astropy.timeseries import BoxLeastSquares
period_grid = np.exp(np.linspace(np.log(1), np.log(15), 50000))
bls = BoxLeastSquares(x, y)
bls_power = bls.power(period_grid, 0.1, oversample=20)
# Save the highest peak as the planet candidate
index = np.argmax(bls_power.power)
bls_period = bls_power.period[index]
bls_t0 = bls_power.transit_time[index]
bls_depth = bls_power.depth[index]
transit_mask = bls.transit_mask(x, bls_period, 0.2, bls_t0)
fig, axes = plt.subplots(2, 1, figsize=(10, 10))
# Plot the periodogram
ax = axes[0]
ax.axvline(np.log10(bls_period), color="C1", lw=5, alpha=0.8)
ax.plot(np.log10(bls_power.period), bls_power.power, "k")
ax.annotate(
"period = {0:.4f} d".format(bls_period),
(0, 1),
xycoords="axes fraction",
xytext=(5, -5),
textcoords="offset points",
va="top",
ha="left",
fontsize=12,
)
ax.set_ylabel("bls power")
ax.set_yticks([])
ax.set_xlim(np.log10(period_grid.min()), np.log10(period_grid.max()))
ax.set_xlabel("log10(period)")
# Plot the folded transit
ax = axes[1]
x_fold = (x - bls_t0 + 0.5 * bls_period) % bls_period - 0.5 * bls_period
m = np.abs(x_fold) < 0.4
ax.plot(x_fold[m], y[m], ".k")
# Overplot the phase binned light curve
bins = np.linspace(-0.41, 0.41, 32)
denom, _ = np.histogram(x_fold, bins)
num, _ = np.histogram(x_fold, bins, weights=y)
denom[num == 0] = 1.0
ax.plot(0.5 * (bins[1:] + bins[:-1]), num / denom, color="C1")
ax.set_xlim(-0.3, 0.3)
ax.set_ylabel("de-trended flux [ppt]")
_ = ax.set_xlabel("time since transit")
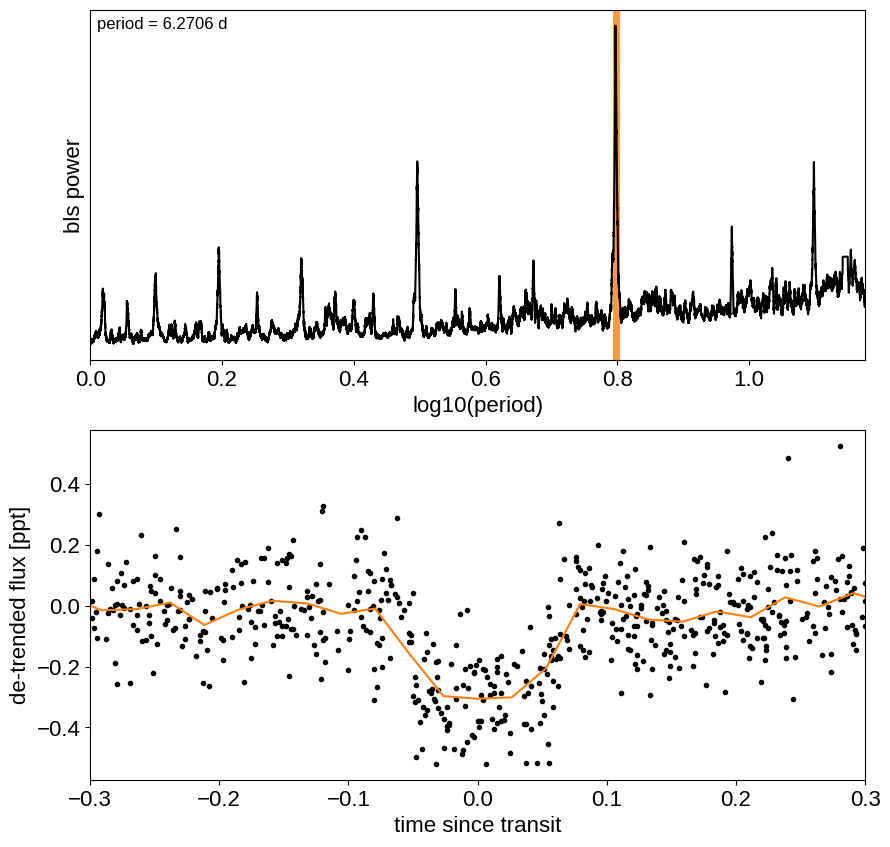
The transit model in PyMC3#
The transit model, initialization, and sampling are all nearly the same as the one in Joint RV & transit fits.
import exoplanet as xo
import pymc3 as pm
import aesara_theano_fallback.tensor as tt
import pymc3_ext as pmx
from celerite2.theano import terms, GaussianProcess
phase_lc = np.linspace(-0.3, 0.3, 100)
def build_model(mask=None, start=None):
if mask is None:
mask = np.ones(len(x), dtype=bool)
with pm.Model() as model:
# Parameters for the stellar properties
mean = pm.Normal("mean", mu=0.0, sd=10.0)
u_star = xo.QuadLimbDark("u_star")
star = xo.LimbDarkLightCurve(u_star)
# Stellar parameters from Huang et al (2018)
M_star_huang = 1.094, 0.039
R_star_huang = 1.10, 0.023
BoundedNormal = pm.Bound(pm.Normal, lower=0, upper=3)
m_star = BoundedNormal(
"m_star", mu=M_star_huang[0], sd=M_star_huang[1]
)
r_star = BoundedNormal(
"r_star", mu=R_star_huang[0], sd=R_star_huang[1]
)
# Orbital parameters for the planets
t0 = pm.Normal("t0", mu=bls_t0, sd=1)
log_period = pm.Normal("log_period", mu=np.log(bls_period), sd=1)
period = pm.Deterministic("period", tt.exp(log_period))
# Fit in terms of transit depth (assuming b<1)
b = pm.Uniform("b", lower=0, upper=1)
log_depth = pm.Normal("log_depth", mu=np.log(bls_depth), sigma=2.0)
ror = pm.Deterministic(
"ror",
star.get_ror_from_approx_transit_depth(
1e-3 * tt.exp(log_depth), b
),
)
r_pl = pm.Deterministic("r_pl", ror * r_star)
# log_r_pl = pm.Normal(
# "log_r_pl",
# sd=1.0,
# mu=0.5 * np.log(1e-3 * np.array(bls_depth))
# + np.log(R_star_huang[0]),
# )
# r_pl = pm.Deterministic("r_pl", tt.exp(log_r_pl))
# ror = pm.Deterministic("ror", r_pl / r_star)
# b = xo.distributions.ImpactParameter("b", ror=ror)
ecs = pmx.UnitDisk("ecs", testval=np.array([0.01, 0.0]))
ecc = pm.Deterministic("ecc", tt.sum(ecs**2))
omega = pm.Deterministic("omega", tt.arctan2(ecs[1], ecs[0]))
xo.eccentricity.kipping13("ecc_prior", fixed=True, observed=ecc)
# Transit jitter & GP parameters
log_sigma_lc = pm.Normal(
"log_sigma_lc", mu=np.log(np.std(y[mask])), sd=10
)
log_rho_gp = pm.Normal("log_rho_gp", mu=0, sd=10)
log_sigma_gp = pm.Normal(
"log_sigma_gp", mu=np.log(np.std(y[mask])), sd=10
)
# Orbit model
orbit = xo.orbits.KeplerianOrbit(
r_star=r_star,
m_star=m_star,
period=period,
t0=t0,
b=b,
ecc=ecc,
omega=omega,
)
# Compute the model light curve
light_curves = (
star.get_light_curve(orbit=orbit, r=r_pl, t=x[mask], texp=texp)
* 1e3
)
light_curve = tt.sum(light_curves, axis=-1) + mean
resid = y[mask] - light_curve
# GP model for the light curve
kernel = terms.SHOTerm(
sigma=tt.exp(log_sigma_gp),
rho=tt.exp(log_rho_gp),
Q=1 / np.sqrt(2),
)
gp = GaussianProcess(kernel, t=x[mask], yerr=tt.exp(log_sigma_lc))
gp.marginal("gp", observed=resid)
# pm.Deterministic("gp_pred", gp.predict(resid))
# Compute and save the phased light curve models
pm.Deterministic(
"lc_pred",
1e3
* star.get_light_curve(
orbit=orbit, r=r_pl, t=t0 + phase_lc, texp=texp
)[..., 0],
)
# Fit for the maximum a posteriori parameters, I've found that I can get
# a better solution by trying different combinations of parameters in turn
if start is None:
start = model.test_point
map_soln = pmx.optimize(
start=start, vars=[log_sigma_lc, log_sigma_gp, log_rho_gp]
)
map_soln = pmx.optimize(start=map_soln, vars=[log_depth])
map_soln = pmx.optimize(start=map_soln, vars=[b])
map_soln = pmx.optimize(start=map_soln, vars=[log_period, t0])
map_soln = pmx.optimize(start=map_soln, vars=[u_star])
map_soln = pmx.optimize(start=map_soln, vars=[log_depth])
map_soln = pmx.optimize(start=map_soln, vars=[b])
map_soln = pmx.optimize(start=map_soln, vars=[ecs])
map_soln = pmx.optimize(start=map_soln, vars=[mean])
map_soln = pmx.optimize(
start=map_soln, vars=[log_sigma_lc, log_sigma_gp, log_rho_gp]
)
map_soln = pmx.optimize(start=map_soln)
extras = dict(
zip(
["light_curves", "gp_pred"],
pmx.eval_in_model([light_curves, gp.predict(resid)], map_soln),
)
)
return model, map_soln, extras
model0, map_soln0, extras0 = build_model()
optimizing logp for variables: [log_rho_gp, log_sigma_gp, log_sigma_lc]
message: Optimization terminated successfully.
logp: 3338.1817955372735 -> 3460.3983411347185
optimizing logp for variables: [log_depth]
message: Optimization terminated successfully.
logp: 3460.3983411347185 -> 3465.961744172654
optimizing logp for variables: [b]
message: Optimization terminated successfully.
logp: 3465.961744172654 -> 3501.4417887424743
optimizing logp for variables: [t0, log_period]
message: Optimization terminated successfully.
logp: 3501.4417887424734 -> 3507.3259047021374
optimizing logp for variables: [u_star]
message: Optimization terminated successfully.
logp: 3507.3259047021384 -> 3510.723293097444
optimizing logp for variables: [log_depth]
message: Optimization terminated successfully.
logp: 3510.723293097444 -> 3514.13173448383
optimizing logp for variables: [b]
message: Optimization terminated successfully.
logp: 3514.13173448383 -> 3515.137731047359
optimizing logp for variables: [ecs]
message: Optimization terminated successfully.
logp: 3515.137731047359 -> 3515.6567965741333
optimizing logp for variables: [mean]
message: Optimization terminated successfully.
logp: 3515.6567965741333 -> 3515.6766245050853
optimizing logp for variables: [log_rho_gp, log_sigma_gp, log_sigma_lc]
message: Desired error not necessarily achieved due to precision loss.
logp: 3515.6766245050853 -> 3516.2740955297063
optimizing logp for variables: [log_sigma_gp, log_rho_gp, log_sigma_lc, ecs, log_depth, b, log_period, t0, r_star, m_star, u_star, mean]
message: Desired error not necessarily achieved due to precision loss.
logp: 3516.274095529706 -> 3722.5360762943724
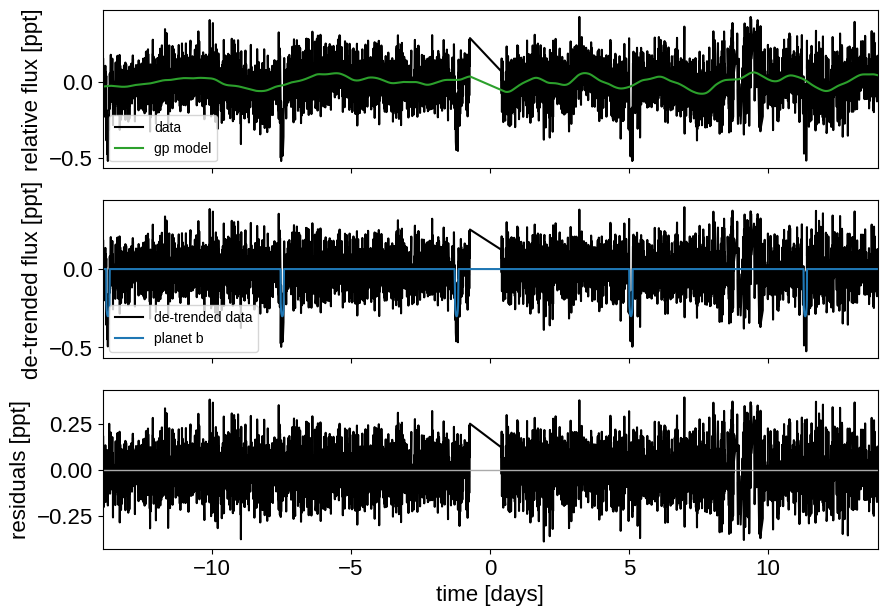
Here’s how we plot the initial light curve model:
def plot_light_curve(soln, extras, mask=None):
if mask is None:
mask = np.ones(len(x), dtype=bool)
fig, axes = plt.subplots(3, 1, figsize=(10, 7), sharex=True)
ax = axes[0]
ax.plot(x[mask], y[mask], "k", label="data")
gp_mod = extras["gp_pred"] + soln["mean"]
ax.plot(x[mask], gp_mod, color="C2", label="gp model")
ax.legend(fontsize=10)
ax.set_ylabel("relative flux [ppt]")
ax = axes[1]
ax.plot(x[mask], y[mask] - gp_mod, "k", label="de-trended data")
for i, l in enumerate("b"):
mod = extras["light_curves"][:, i]
ax.plot(x[mask], mod, label="planet {0}".format(l))
ax.legend(fontsize=10, loc=3)
ax.set_ylabel("de-trended flux [ppt]")
ax = axes[2]
mod = gp_mod + np.sum(extras["light_curves"], axis=-1)
ax.plot(x[mask], y[mask] - mod, "k")
ax.axhline(0, color="#aaaaaa", lw=1)
ax.set_ylabel("residuals [ppt]")
ax.set_xlim(x[mask].min(), x[mask].max())
ax.set_xlabel("time [days]")
return fig
_ = plot_light_curve(map_soln0, extras0)
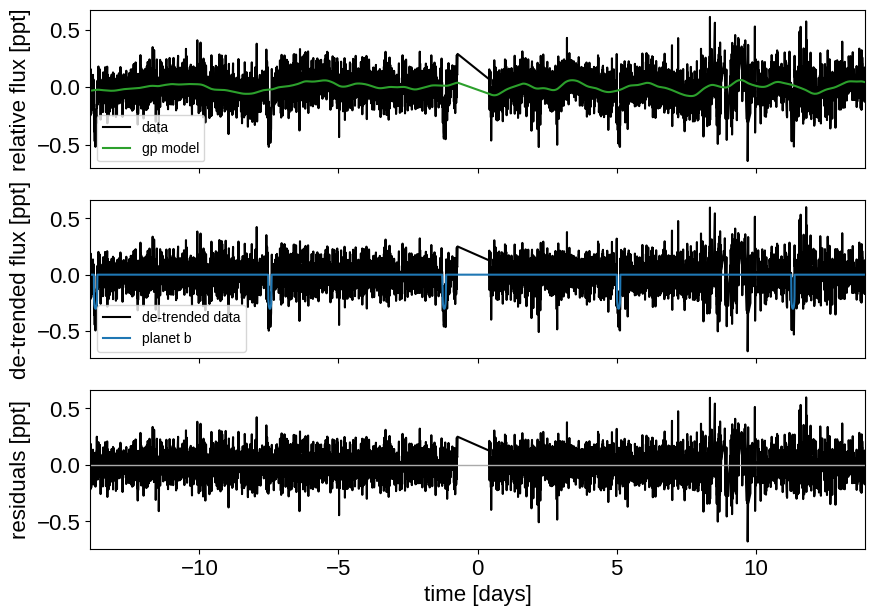
As in Joint RV & transit fits, we can do some sigma clipping to remove significant outliers.
mod = (
extras0["gp_pred"]
+ map_soln0["mean"]
+ np.sum(extras0["light_curves"], axis=-1)
)
resid = y - mod
rms = np.sqrt(np.median(resid**2))
mask = np.abs(resid) < 5 * rms
plt.figure(figsize=(10, 5))
plt.plot(x, resid, "k", label="data")
plt.plot(x[~mask], resid[~mask], "xr", label="outliers")
plt.axhline(0, color="#aaaaaa", lw=1)
plt.ylabel("residuals [ppt]")
plt.xlabel("time [days]")
plt.legend(fontsize=12, loc=3)
_ = plt.xlim(x.min(), x.max())
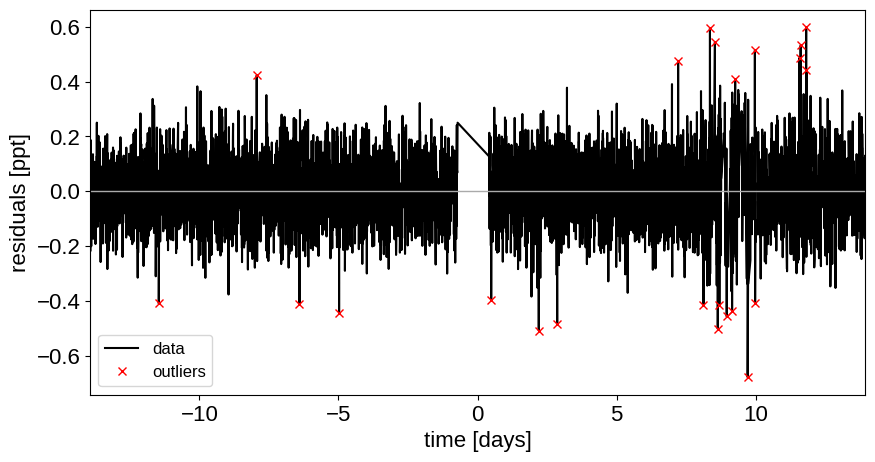
And then we re-build the model using the data without outliers.
model, map_soln, extras = build_model(mask, map_soln0)
_ = plot_light_curve(map_soln, extras, mask)
optimizing logp for variables: [log_rho_gp, log_sigma_gp, log_sigma_lc]
message: Optimization terminated successfully.
logp: 3880.262483379479 -> 3886.439828473729
optimizing logp for variables: [log_depth]
message: Optimization terminated successfully.
logp: 3886.439828473729 -> 3886.4414819670956
optimizing logp for variables: [b]
message: Optimization terminated successfully.
logp: 3886.4414819670956 -> 3886.441650516504
optimizing logp for variables: [t0, log_period]
message: Desired error not necessarily achieved due to precision loss.
logp: 3886.4416505165045 -> 3886.4418199582783
optimizing logp for variables: [u_star]
message: Optimization terminated successfully.
logp: 3886.441819958278 -> 3886.441904696529
optimizing logp for variables: [log_depth]
message: Optimization terminated successfully.
logp: 3886.441904696529 -> 3886.441910233041
optimizing logp for variables: [b]
message: Optimization terminated successfully.
logp: 3886.441910233041 -> 3886.4419208006507
optimizing logp for variables: [ecs]
message: Optimization terminated successfully.
logp: 3886.44192080065 -> 3886.4419209254083
optimizing logp for variables: [mean]
message: Optimization terminated successfully.
logp: 3886.4419209254097 -> 3886.444210713328
optimizing logp for variables: [log_rho_gp, log_sigma_gp, log_sigma_lc]
message: Optimization terminated successfully.
logp: 3886.444210713328 -> 3886.444220152795
optimizing logp for variables: [log_sigma_gp, log_rho_gp, log_sigma_lc, ecs, log_depth, b, log_period, t0, r_star, m_star, u_star, mean]
message: Desired error not necessarily achieved due to precision loss.
logp: 3886.444220152795 -> 3886.4442436117993
Now that we have the model, we can sample:
import platform
with model:
trace = pm.sample(
tune=1500,
draws=1000,
start=map_soln,
# Parallel sampling runs poorly or crashes on macos
cores=1 if platform.system() == "Darwin" else 2,
chains=2,
target_accept=0.95,
return_inferencedata=True,
random_seed=[261136679, 261136680],
init="adapt_full",
)
Auto-assigning NUTS sampler...
Initializing NUTS using adapt_full...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [log_sigma_gp, log_rho_gp, log_sigma_lc, ecs, log_depth, b, log_period, t0, r_star, m_star, u_star, mean]
Sampling 2 chains for 1_500 tune and 1_000 draw iterations (3_000 + 2_000 draws total) took 513 seconds.
import arviz as az
az.summary(
trace,
var_names=[
"omega",
"ecc",
"r_pl",
"b",
"t0",
"period",
"r_star",
"m_star",
"u_star",
"mean",
],
)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| omega | 0.450 | 1.785 | -2.817 | 3.129 | 0.056 | 0.040 | 1139.0 | 1690.0 | 1.0 |
| ecc | 0.221 | 0.147 | 0.000 | 0.485 | 0.006 | 0.005 | 720.0 | 758.0 | 1.0 |
| r_pl | 0.018 | 0.001 | 0.017 | 0.020 | 0.000 | 0.000 | 1196.0 | 1384.0 | 1.0 |
| b | 0.455 | 0.222 | 0.028 | 0.773 | 0.009 | 0.007 | 573.0 | 630.0 | 1.0 |
| t0 | -13.734 | 0.002 | -13.739 | -13.731 | 0.000 | 0.000 | 1333.0 | 1412.0 | 1.0 |
| period | 6.269 | 0.001 | 6.267 | 6.270 | 0.000 | 0.000 | 1703.0 | 1527.0 | 1.0 |
| r_star | 1.100 | 0.023 | 1.057 | 1.143 | 0.001 | 0.000 | 1927.0 | 1367.0 | 1.0 |
| m_star | 1.097 | 0.038 | 1.024 | 1.167 | 0.001 | 0.001 | 1826.0 | 1486.0 | 1.0 |
| u_star[0] | 0.302 | 0.227 | 0.000 | 0.704 | 0.006 | 0.004 | 1466.0 | 1054.0 | 1.0 |
| u_star[1] | 0.219 | 0.307 | -0.304 | 0.807 | 0.008 | 0.006 | 1319.0 | 1162.0 | 1.0 |
| mean | 0.002 | 0.007 | -0.012 | 0.016 | 0.000 | 0.000 | 1699.0 | 1098.0 | 1.0 |
Results#
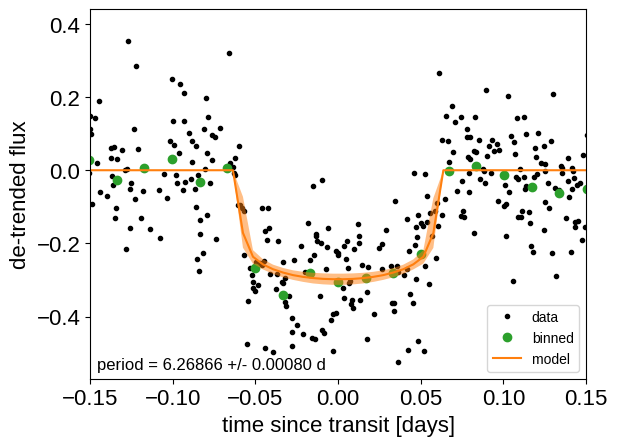
After sampling, we can make the usual plots. First, let’s look at the folded light curve plot:
flat_samps = trace.posterior.stack(sample=("chain", "draw"))
# Compute the GP prediction
gp_mod = extras["gp_pred"] + map_soln["mean"] # np.median(
# flat_samps["gp_pred"].values + flat_samps["mean"].values[None, :], axis=-1
# )
# Get the posterior median orbital parameters
p = np.median(flat_samps["period"])
t0 = np.median(flat_samps["t0"])
# Plot the folded data
x_fold = (x[mask] - t0 + 0.5 * p) % p - 0.5 * p
plt.plot(x_fold, y[mask] - gp_mod, ".k", label="data", zorder=-1000)
# Overplot the phase binned light curve
bins = np.linspace(-0.41, 0.41, 50)
denom, _ = np.histogram(x_fold, bins)
num, _ = np.histogram(x_fold, bins, weights=y[mask])
denom[num == 0] = 1.0
plt.plot(
0.5 * (bins[1:] + bins[:-1]), num / denom, "o", color="C2", label="binned"
)
# Plot the folded model
pred = np.percentile(flat_samps["lc_pred"], [16, 50, 84], axis=-1)
plt.plot(phase_lc, pred[1], color="C1", label="model")
art = plt.fill_between(
phase_lc, pred[0], pred[2], color="C1", alpha=0.5, zorder=1000
)
art.set_edgecolor("none")
# Annotate the plot with the planet's period
txt = "period = {0:.5f} +/- {1:.5f} d".format(
np.mean(flat_samps["period"].values), np.std(flat_samps["period"].values)
)
plt.annotate(
txt,
(0, 0),
xycoords="axes fraction",
xytext=(5, 5),
textcoords="offset points",
ha="left",
va="bottom",
fontsize=12,
)
plt.legend(fontsize=10, loc=4)
plt.xlim(-0.5 * p, 0.5 * p)
plt.xlabel("time since transit [days]")
plt.ylabel("de-trended flux")
_ = plt.xlim(-0.15, 0.15)
And a corner plot of some of the key parameters:
import corner
import astropy.units as u
trace.posterior["r_earth"] = (
trace.posterior["r_pl"].coords,
(trace.posterior["r_pl"].values * u.R_sun).to(u.R_earth).value,
)
_ = corner.corner(
trace,
var_names=["period", "r_earth", "b", "ecc"],
labels=[
"period [days]",
"radius [Earth radii]",
"impact param",
"eccentricity",
],
)
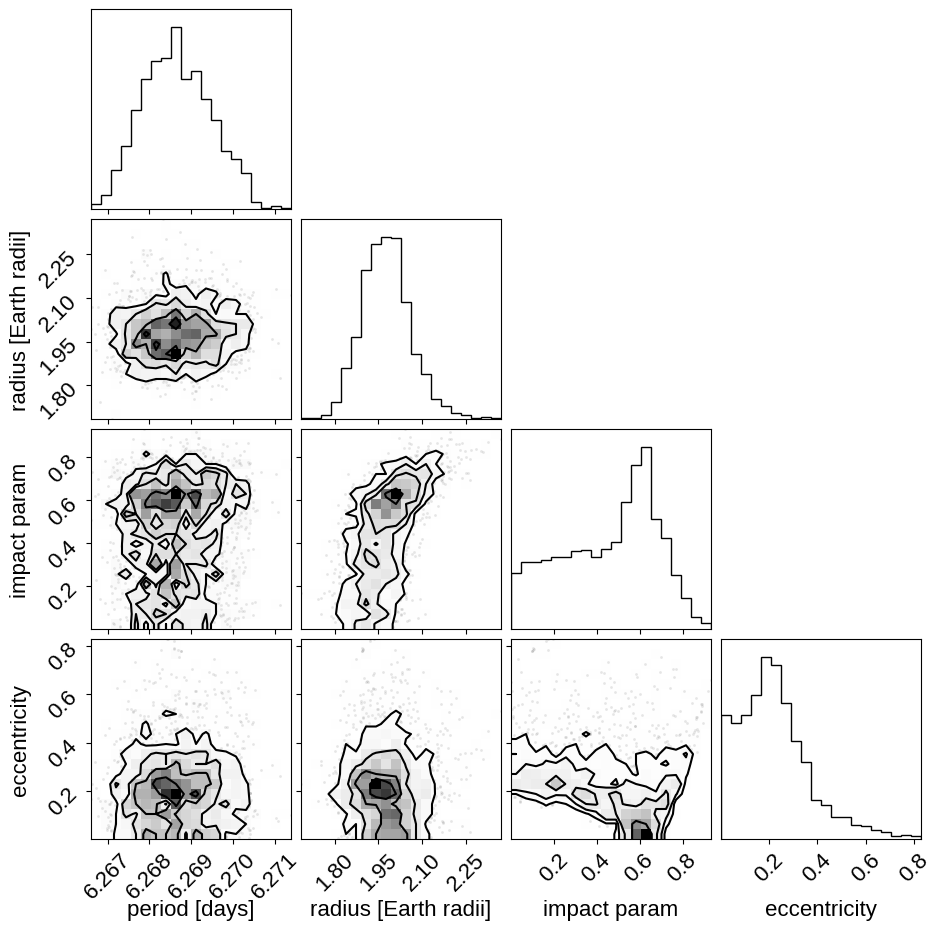
These all seem consistent with the previously published values.
Citations#
As described in the citation tutorial, we can use citations.get_citations_for_model to construct an acknowledgement and BibTeX listing that includes the relevant citations for this model.
with model:
txt, bib = xo.citations.get_citations_for_model()
print(txt)
This research made use of \textsf{exoplanet} \citep{exoplanet:joss,
exoplanet:zenodo} and its dependencies \citep{celerite2:foremanmackey17,
celerite2:foremanmackey18, exoplanet:agol20, exoplanet:arviz,
exoplanet:astropy13, exoplanet:astropy18, exoplanet:kipping13,
exoplanet:kipping13b, exoplanet:luger18, exoplanet:pymc3, exoplanet:theano}.
print(bib.split("\n\n")[0] + "\n\n...")
@article{exoplanet:joss,
author = {{Foreman-Mackey}, Daniel and {Luger}, Rodrigo and {Agol}, Eric
and {Barclay}, Thomas and {Bouma}, Luke G. and {Brandt},
Timothy D. and {Czekala}, Ian and {David}, Trevor J. and
{Dong}, Jiayin and {Gilbert}, Emily A. and {Gordon}, Tyler A.
and {Hedges}, Christina and {Hey}, Daniel R. and {Morris},
Brett M. and {Price-Whelan}, Adrian M. and {Savel}, Arjun B.},
title = "{exoplanet: Gradient-based probabilistic inference for
exoplanet data \& other astronomical time series}",
journal = {arXiv e-prints},
year = 2021,
month = may,
eid = {arXiv:2105.01994},
pages = {arXiv:2105.01994},
archivePrefix = {arXiv},
eprint = {2105.01994},
primaryClass = {astro-ph.IM},
adsurl = {https://ui.adsabs.harvard.edu/abs/2021arXiv210501994F},
adsnote = {Provided by the SAO/NASA Astrophysics Data System}
}
...

